Article Summary 25/09/21
Article title
Diffusing protein binders to intrinsically disordered proteins
Journal
Nature
Tags
Protein design; RFdiffusion; Protein tool development
Introduction
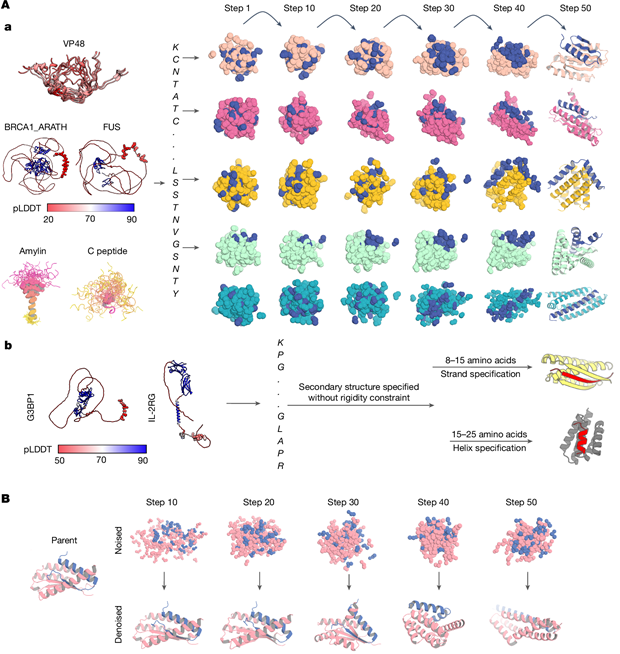
Intrinsically disordered proteins represent ~50% of human proteome. It exerts biological functions without specific secondary structure of peptide. IDPs act as vital effector in disease generation, like amylin in diabetes, and C-peptide as biomarker for diabetes.
Therefore, designing IDP binders helps to understand and regulate such disease and related pathways. Hitherto, binder design is only limited to structured target, while binders of unstructured targets such as IDPs mainly derived from high-cost and low-reproducibility antibody production. Protein computational design towards unstructured targets is urgently needed.
This work
Principle
Specifically-trained RFdiffusion model from 2-chain PDB database (1 chain structure given, 1 chain only sequence given, structure randomized) to design proteins. In training only 50% of complete noise time is consumed, resulting in limited geometry with some randomness, rather than totally noised. Therefore using this trained model, we could input only target sequence to design a binder towards this target. In design, both target and binder is noised and denoised, which is different from previous approach in which only binder is noised and denoised. Thus the strategy is called two-sided partial diffusion.
For short unstructured target peptides: also pretrained RFdiffusion model, capable of annotating specified secondary structures at a target region. Through this strategy, we can design target-binder pair other than helix-based interactions (maintained through hydrophobic interactions) which are prevalent in protein design, diversifying the designed binder pool.
Validation
In silicon validation: Rosetta relax (pLDDT; ddG etc.) to screen ~100 designs from ~10-50k candidates
Binding evaluation: most binders bind target at 3-100 nM level.
Crystal structure: RMSD of αC of predicted-resolved is low enough to confirm the correct design of binder-target interactions.
Cellular validation: binders bind target in cell and affect physical properties of targets in cell.
Disease-related: binder prevents amylin aggregation in cell and tag-fused binder cause amylin transported to lysozyme to degradation.
doi
10.1038/s41586-025-09248-9