Article-summary-03-07-25
Article title
A PARP2 active site helix melts to permit DNA damage-induced enzymatic activation
Journal
Molecular Cell
Tags
ADP-ribosylation; Allosteric activation; HDX-MS
Introduction
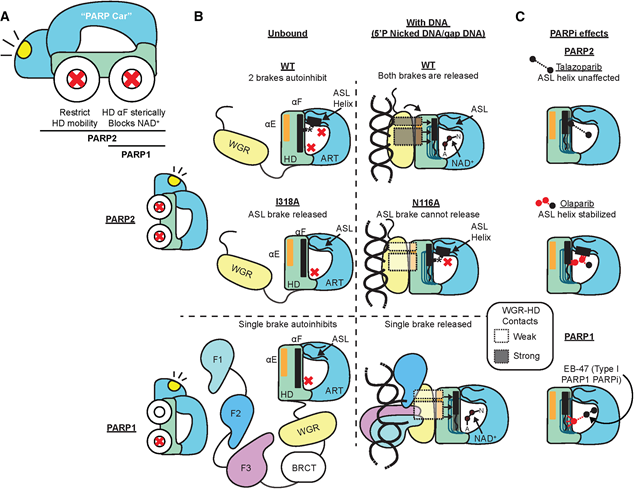
PARP1 and PARP2, both ADP-ribosylation writer, regulate DNA damage response and act as attractive drug target. In apo state, PARP1 and PARP2 show auto-inhibition, as DNA binding allosterically activates them to perform ADP-ribosylation activity. Previous studies indicated PARP1 is activated by HD unfolding upon DNA recognition. However, whether PARP2 adapts similar activation mechanism compared to PARP1 remains elusive.
This work
The article utilized HDX-MS to probe the conformational change of PARP2 upon DNA binding. Through this approach, this article finds a new helix ASL in PARP2 regulates PARP2 activation, showing distinct activation pattern compared to PARP1.
First, the article found that not only HD, but also ASL underwent DNA recognition-mediated HX signal change, indicating ASL played role in DNA-mediated PARP2 allosteric activation. They found a PARP2 mutant N116A that kept DNA binding and HD unfolding feature, but lost the ability of catalysis. In N116A mutant, ASL helix is not unfolded upon DNA binding, implying that PARP2 ASL helix unfolding is also an essential prior qualification for PARP2 activation. Moreover, they found HD domain interacts with ASL, stabilizing its helix to inhibit PARP2. The article mainly uses HDX-MS technology, with scarce biochemical confirmation. The importance of this article is to claim the divergence of allosteric activation mode between PARP1 and PARP2.
doi
doi.org/10.1016/j.molcel.2025.01.004