Article summary 8/21/24
Article title
A ubiquitin-specific, proximity-based labeling approach for the identification of ubiquitin ligase substrates
Journal
Science advances
Tags
E3 ligase; Promixity labeling
Introduction
Dissecting E3 ligase substrates is of great importance for studying its cellular function. However, current methodologies could not target a specific E3 (di-GG antibody), or disturbs the ubiquitination pathway (biotin-Ub; biotin-Ub-E3 fusion). Thus, alternative E3 substrate screening approach is needed to expand the substrate screening toolbox.
This work
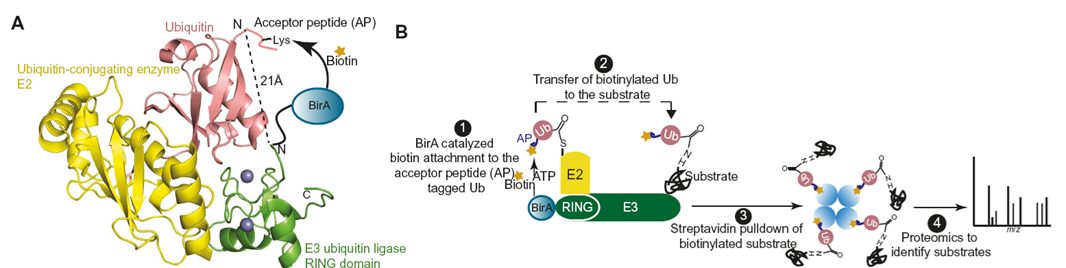
Develop a strategy called Ub-POD (proximity and orientation dependent tagging of Ub). With a BirA fused to target E3, AP-tagged Ub is biotinylated and attached to substrate. biotinylated and ubiquitinated substrate are enriched and MS analyzed.
RAD18 is used as a model system to probe E3 substrate. In addition to optimize Ub-POD, they edited the AP tag to regulate BirA-Ub affinity and inspect its spatial specificity. Then other 2 E3 ligases (CHIP and TRAF6) are applied to screen their substrates.